CellMarkerPipe cell marker identification and evaluation pipeline in single cell transcriptomes
Published onDecember 31, 2023
-Views
1Minutes Read
My Research Publications
CellMarkerPipe: cell marker identification and evaluation pipeline in single cell transcriptomes
Authors: Yinglu Jia, Pengchong Ma, Qiuming Yao
Published in: Scientific Reports, 14, 13151 (2024)
Abstract:
Assessing marker genes from all cell clusters can be time-consuming and lack systematic strategy. Streamlining this process through a unified computational platform that automates identification and benchmarking will greatly enhance efficiency and ensure a fair evaluation. We therefore developed a novel computational platform, cellMarkerPipe (https://github.com/yao-laboratory/cellMarkerPipe), for automated cell-type specific marker gene identification from scRNA-seq data, coupled with comprehensive evaluation schema. CellMarkerPipe adaptively wraps around a collection of commonly used and state-of-the-art tools, including Seurat, COSG, SC3, SCMarker, COMET, and scGeneFit. From rigorously testing across diverse samples, we ascertain SCMarker’s overall reliable performance in single marker gene selection, with COSG showing commendable speed and comparable efficacy. Furthermore, we demonstrate the pivotal role of our approach in real-world medical datasets. This general and opensource pipeline stands as a significant advancement in streamlining cell marker gene identification and evaluation, fitting broad applications in the field of cellular biology and medical research.
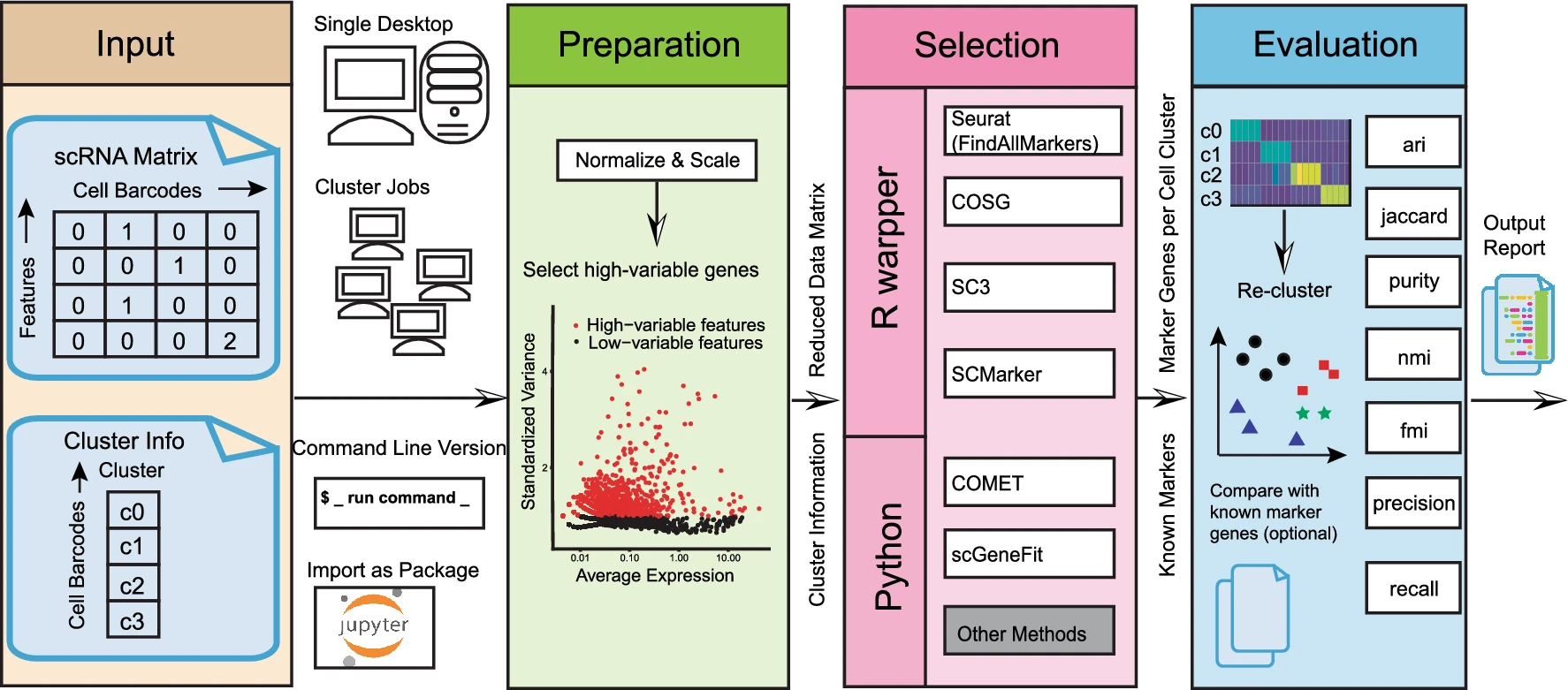
How to Use
- Cite This Article: Jia, Y., Ma, P. & Yao, Q. CellMarkerPipe: cell marker identification and evaluation pipeline in single cell transcriptomes. Sci Rep 14, 13151 (2024). https://doi.org/10.1038/s41598-024-63492-z.
- Read the Full Paper: Access Here
Tags:
#Single cell